PhD thesis committee meeting
Shaun Jackman
2018-03-14
Shaun Jackman
Thesis Committee
Timeline
- Previous meeting 2015-07-27
- Fourth committee meeting 2018-03-14
- Start writing early 2018
- Submit thesis and defend late 2018
- Graduate spring 2019
Conferences
- Three talks and four posters
- ISMB 2016 BOSC talk and poster
Linuxbrew and Homebrew for Cross-platform Package Management - ISMB 2016 HiTSeq poster
Organellar Genomes of White spruce (Picea glauca): Assembly and annotation - PAG 2017 talk and AGBT 2017 poster
Organellar Genomes of Sitka Spruce (Picea sitchensis): Assembly and Annotation - ISMB 2017 talk and poster
Tigmint: Correct Misassemblies Using Linked Reads From Large Molecules
Teaching Assistant
UBC Master of Data Science
Papers
- Assembly of the complete Sitka spruce chloroplast genome using 10X Genomics’ GemCode sequencing data L Coombe, RL Warren, SD Jackman, C Yang, BP Vandervalk, RA Moore, …
PloS one 2016 - Spaced seed data structures for de novo assembly
I Birol, J Chu, H Mohamadi, SD Jackman, K Raghavan, BP Vandervalk, …
International journal of genomics 2015 - Konnector v2.0: pseudo-long reads from paired-end sequencing data
BP Vandervalk, C Yang, Z Xue, K Raghavan, J Chu, H Mohamadi, SD Jackman,…
BMC medical genomics 2015 - Sealer: a scalable gap-closing application…
D Paulino, RL Warren, BP Vandervalk, A Raymond, SD Jackman, I Birol
BMC Bioinformatics 2015 - Improved white spruce (Picea glauca) genome…
RL Warren, CI Keeling, MMS Yuen, A Raymond, GA Taylor, …
The Plant Journal 2015
Papers
- Three first-author (or joint) papers since 2015
- Collaborated on 29 papers since 2009
- 25 papers with at least 10 citations
- Two first-author manuscripts in preparation
Manuscripts
- ORCA: A Comprehensive Bioinformatics Container Environment for Education and Research SD Jackman*, T Mozgacheva*, B O’Huiginn, L Bailey, I Birol, SJM Jones
- Tigmint: Correct Assembly Errors Using Linked Reads From Large Molecules SD Jackman, J Chu, RL Warren, BP Vandervalk, L Coombe, S Yeo, …
Thesis Outline
Methods
- ABySS 2.0: resource-efficient assembly of large genomes
- Tigmint: Correcting misassemblies using linked reads
- UniqTag: Content-derived unique and stable identifiers
Genomes
- Organellar genomes of white spruce (Picea glauca)
using paired-end and mate-pair reads - Organellar genomes of Sitka spruce (Picea sitchensis)
using linked reads and Nanopore reads - Genome assembly of western redcedar (Thuja plicata)
using paired-end, mate-pair, and linked reads
White Spruce Organelles
- Assembled cpDNA and mtDNA genomes
- Annotated genes (mRNA, rRNA, tRNA) and repeats
- Analysed RNA-seq data to quantify
- transcript abundance in eight tissues
- expressed ORFs
- C-to-U RNA editing
- cryptic ACG start codons due to C-to-U RNA editing
- Submitted annotated genomes to GenBank
- Published paper in Genome Biology and Evolution (2015)

Sitka Spruce Plastid
- Assembled by Rene Warren and Lauren Coombe
using linked reads - Annotated genes (mRNA, rRNA, tRNA)
- Submitted annotated genomes to GenBank
- Published paper in PLOS ONE (2016)

Sitka Spruce Mitochondrion
- 10x Genomics Chromium sequencing
- > 50x mitochondrial coverage in one lane
- 11 lanes of Oxford Nanopore Sequencing
- 3x nuclear coverage
- 14x mitochondrial coverage
- Assemble Nanopore reads
with Canu and Unicycler - Scaffold and polish with linked reads

Sitka Spruce Mitochondrion
- Complete the genome assembly
- Determine chromosomal structure
- Annotate the genome
- Submit the genome to GenBank
- Write the manuscript
ABySS 2.0
- Implemented Bloom filter de Bruijn Graph
- Konnector and Konnector 2.0 with Ben Vandervalk
- Sealer with Daniel Paulino
- ABySS 2.0 with Ben Vandervalk
- Reduce memory usage by twelve fold over ABySS 1.0
- Assemble a conifer genome with a single machine
- Memory usage is independent of parameter k
- Assembled a human genome with ABySS 2.0 (35 GB RAM)
ABySS 2.0
- Compared to ABySS 1.5 and six other assemblers
- Submitted genome assemblies to NCBI
- Published paper in Genome Research (2017)
- Presenting a talk at RECOMB-Seq 2018
Tigmint
Correcting misassemblies using linked reads
- Incorrectly assembled sequence complicates all downstream analyses
- Misassemblies also limit contiguity
- Cut contigs where linked reads and assembly disagree
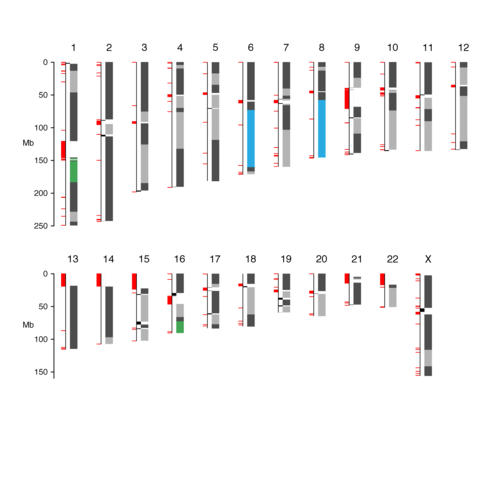
- Tigmint + ARCS improved contiguity two fold over ARCS alone in human from 8 Mbp to 16 Mbp scaffold NGA50
- Further developed the tool with Lauren Coombe
- Presenting a talk and poster at RECOMB-Seq 2018


Western Redcedar (Thuja plicata)
Method
- Trim adapters with Trimadap and NxTrim
- Count k-mers with ntCard
- Estimate genome size GenomeScope
- Assemble PE and MP reads with ABySS 2.0
- Scaffold with Chromium reads using ARCS
- Assess genome completeness using BUSCO
Results
- Scaffold NG50 of 1.2 Mbp
- BUSCO 54% complete, 7% fragmented
Thesis Outline
Methods
- ABySS 2.0: resource-efficient assembly of large genomes
- Tigmint: Correcting misassemblies using linked reads
- UniqTag: Content-derived unique and stable identifiers
Genomes
- Organellar genomes of white spruce (Picea glauca)
using paired-end and mate-pair reads - Organellar genomes of Sitka spruce (Picea sitchensis)
using linked reads and Nanopore reads - Genome assembly of western redcedar (Thuja plicata)
using paired-end, mate-pair, and linked reads